Was the garre ev32? What clan was the rx and what's his haplogroup?Yes. If you meant « which tribe do you mostly match » I’d say Isaaq and Dir. my top 40 matches seem to all come from (or partially) western and central SL, and it makes sense since I’m HA. The other matches are quite irrelevant since they’re pretty distant. The rest of my 800 matches are from northeast, central Somalia and Galbeed. I think I matched with very little southern Somalis.
It seems like the divide between « Sab and Samaale » clans is a real thing.
I’ve spoken to a RX brother and Garre one, both told me that they have just a couple hundred matches. Bare in mind, your average northern/central Somali person gets around 700-1000 matches on there. Quite interesting.
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
GENETICS Y-DNA and mtDNA of Somali (Mostly) 23andMe Relatives
- Thread starter Nabiil
- Start date
The alchemist
VIP
The latest manual labor results a while back before I found out there was a download button:E-V32 -> 72.65%
T-L208 & T-M70 -> 15.59% & 1.18% = 16.77%
E-V32 = 60%
T-L208 = 24.2%
T-M70 = 1.57%
The alchemist
VIP
I could have taken a representative measure instead of going through what then was everything. Still, I checked each available because I did not want to miss out on rare haplogroups. I thought checking for autosomal peculiarities was convenient since I was going through all the work anyway.
They disabled it? Thankfully I downloaded my stuff.
Dude, that number I showed was all done through manual labor. Sifting through 100s of people was tiresome and tedious.
I could have taken a representative measure instead of going through what then was everything. Still, I checked each available because I did not want to miss out on rare haplogroups. I thought checking for autosomal peculiarities was convenient since I was going through all the work anyway.
You don't have permission to view the spoiler content.
Log in or register now.
I took the manual approach on only the non-E-V32 and non-T Y-DNA haplogroups, around a dozen of them were false matches/recent mixes, so I just filtered those out easy enough. The task is more tedious for mtdna, my relatives list has 120 different rows of mtdna haplogroups, so I didn't bother with those. Some are obvious but others with M, N, L3 are too similar.
The alchemist
VIP
Why do they duplicate the results of some people on the downloaded files?You don't have permission to view the spoiler content. Log in or register now.
I took the manual approach on only the non-E-V32 and non-T Y-DNA haplogroups, around a dozen of them were false matches/recent mixes, so I just filtered those out easy enough. The task is more tedious for mtdna, my relatives list has 120 different rows of mtdna haplogroups, so I didn't bother with those. Some are obvious but others with M, N, L3 are too similar.
Initially I thought it was a fluke, then tried downloading for a second time and often I see same people double or triple in a row on the spreadsheet.
They list every chromosomal segment you share with each person in separate rows. I removed the duplicates based on the profile url column which stays the same for each person; whether they only share 1 chromosomal segment (1 row) or they're your parent who shares 23 segments (23 rows), rather than removing duplicates by name and risking losing people with commonly shared names.Why do they duplicate the results of some people on the downloaded files?
View attachment 314219
Initially I thought it was a fluke, then tried downloading for a second time and often I see same people double or triple in a row on the spreadsheet.
@The alchemist @Garaad Awal
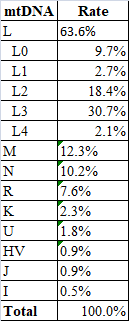
mtDNA from 727 relatives
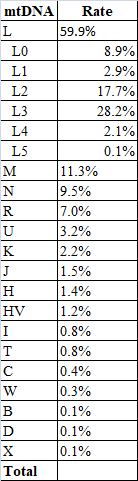
After taking out non-Somali maternal lineages, N=659.
mtDNA from 727 relatives

After taking out non-Somali maternal lineages, N=659.
You don't have permission to view the spoiler content.
Log in or register now.

Last edited:
The alchemist
VIP
Here is my list from a while back (mixed people were removed):@The alchemist @Garaad Awal
mtDNA from 727 relatives

After taking out non-Somali maternal lineages, N=659.
You don't have permission to view the spoiler content. Log in or register now.

I have seen others post results. It seems consistent. It would be fair to recognize some bias in our pooling due to overlapping sample sets.
Nevertheless, I will state these values to be very representative of macro northern Somali mtDNA with some minor adjustments proven through the fact that you have nearly 2x my sample size without significant deviation. The positive thing is the sample bias is geared toward Af Maxaa, so we should suspect all the large clans to have this. Well, I don't know about Maay speakers, even if you control mixing -- it could be that mtDNA founder effects have shaped different pockets of clusters from the pool we listed. Or it could be that we have received bottle-neck where diversity has lowered and retained in southern Somalia. These things are not mutually exclusive. We will see. Southern Somalians need a study of its own to cover this, in my opinion.
v coolHello there,
Through google search I came across this topic and I thought I might interest you with my findings as an Eritrean with roots in the central province (maternally and paternally).
View attachment 243857
Sample size = 54 (shared data on 23andMe)
View attachment 243858
Sample size = 147
Most common haplogroups pairs: A1b1-M118 with M1a1; E-M35 with L2/L3; J with U
----
A1b1-M118 (nilotic clade) ~10kyears (tmrca)
M1a1 (north east africa) ~13kyears
----
E-M35 (north africa/northern horn of africa) ~22kyears
L2 and L3 clades (african clades) ~80kyears
----
J clade (anatolia/caucasus) ~30k years
U clade (west asia/caucasus) ~40k years
View attachment 243859
Sample size = 68
Most common L2 clade = L2a1 by 77,3% towards other L2 clades
Most common L3 clades = L3f by 43,8% and L3i by 21,9% towards other L3 clades.
Personal haplogroup E-V32 and L2a1
My terminal haplogroup falls within saudis on ftdna unlike most somalis
I have a closer common haplogroup with somalis with whom I share a common ancestor dating back to 3000 BCE, but i'm closer with saudis with whom I share a common ancestor dating back ~500 BCE. As if my paternal ancestors had only recently migrated in or out (my bet is on the migration in from arabia after clashes with jewish tribes 1500 years ago since I'm of a christian orthodox descent).
I just used Dnagenics matches analyses tool and uploaded the relatives data thereWhy do they duplicate the results of some people on the downloaded files?
View attachment 314219
Initially I thought it was a fluke, then tried downloading for a second time and often I see same people double or triple in a row on the spreadsheet.
You don't have permission to view the spoiler content.
Log in or register now.
Log in - DNA Genics
www.dnagenics.com
Trending
-
-
Myron Gaines Glazes A White Nationalist Just To Get Called "A Based N*gger" 💀
- Started by _Yusuf_
- Replies: 43
-
