@Asrafael, @embarassing, @Samaalic Era, @Apollo, @Alexis
Somalispot Department of Suugo Science
Ministry of Dila, Hala Dilo, Somaali Ma Aha
@Apollo Medical School, Institute for Cilmi Bariis
For weeks, the Somalispot community has been plagued with a trail of unfettered excreta of unidentified provenance coursing it's way through the sub-forum and forum at large. Several members of the community have taken at attempts in trying to source the origination of the perceivingly reproductive excreta. The characteristics of the subject excreta included, among several, a malodorous smell, elevated levels of hemoglobin blood iron, and plasma seemingly sourced from a variety of ruminants, and a tendency to proliferate due to unknown and yet uninvestigated means, and a symbiotic relationship with lice larvae and colonia.
The unbefore heard of set of traits that characterize the subject excreta have led to an abundance of theories to explain its source of origination. The following are a compilation of multiple theories put forth by several individuals and community members :
1) the Italian origination theory, which posits its substance on the rather unique Italian sourced name used by a user who is believed by some to be well timed and placed in association with this phenomena, going off the connection between Farabutto -> Farabuuto. This is despite it's usage amongst few Somalis.
2) the Eritrean origination theory, due to a battery if 15N and 13C- isotope analysis and dietary values of the subject excreta all aligning with the Tigray serie, consistent with its latent opposition to Amara and Galla sample sets, and the results of mass spectrometry having a positive association with previous collected Eritrean results
3) the Waaqo theory, due to the subjects excreta rather vague resemblant character traits
In addition to these three, there exists a forth theory, the hybrid theory, which characterized by the idea in which theories 2 and 3 having to not be mutually exclusive theories, but are in fact evidence of each other, based off a premise that waaqo was Eritrean, and not a non-Eritrean injirleey.
As of yet, the subject excreta has yet to have been identified with any association with Somalis, but by testing the probable occurrence of the latter theory, we found
O (x | y), with O (waaqo) = .30, and
O (waaqo | similarity of posts ) = .100
Which supports the Waaqo theory as having a higher probability of occurring with the known and considered probability factors.
In combination with our 13^C and 15^N isotope analysis, mass spectrometry, and dietary values all suggesting an Eritrean origin for the originating source of the subject excreta, independent of genetic evidence, we suggest with strong confidence in the hybrid theory, and for an Eritrean background for waaqoofpunt, and waaqoofpunt as the source of the offending subject excreta.
But due to the unsatisfactory and lack of robustness of the evidence thus given, the rather harsh argumentative and currently subjective nature of the discussion, and the uncooperative and fragile nature of the subject excreta, we found it necessary to perform a genetic analysis.
We genotyped samples which were collected with the extraction of a acidic core located on a remnant stain of the subject excreta which was received from @embarassing. We then prepared the sampled core, and successfully searched and extracted biopolymers, and using an extraction buffer consisting of proteinase k and EDTA and any needed decalsification, we estimated a yield of near 100% after centrifuge, and the ensuing yield was put in a polymerase chain reaction with several identical copies. We used capillary electrophoresis in the separation process, and identified all with a high power of discrimination all locus of interest in regards to the subject excreta. The MWF of our yield had a A260-80 ratio 1.9. We genotyped a total of 500k SNPs, and compared them to collated set of genotyped excreta remains and a collection of various African populations, with Han, Maya, Injirleey, san-Gaab, and Somali results, the latter and injirleey samples were collected being local community members. In order to test for the waaqo theory, we asked and received excrement collected and stored by the administration for purposes of identification of the two time banned member @waaqoofpunt. We similarly received further samples provided by community members of the same purported user found to be on twitter. We then pruned the collected set for any significant outliers that could hamper and mischarecterize our analysis, and had to remove the following samples for the following reasons:
@CaliTedesse (self identified Arabian, excess Somali ancestry)
@Samaalic Era (extremely high RoH)
@Medulla (unstable and erratic mutation rate).
@Basra (self identified as female, genotyped as XY)
We collated and integrated the Somali samples into a single high density sample in order to have a very high level of snp recall >99%.
By F-stats, we found a very high allele sharing between the subject excreta and the sole injirleey, and running F4 and F3 with Han, san-Gaab, and Chimp outgroup, see strong support for a common node including both injirleey and the subject excreta.
F4 was run as given: F4 (injirleey, subjectexcreta | san-Gaab, Han) = 0
thus supporting a common node and subset including injirleey, subjectexcreta.
We then ran F4 I with Somali, sanGaab to see a further common branching for this node
F4 (injirleey, subjectexcreta | Somali, sanGaab) was null, which establishes a very strong basis for a special relationship of injirleey and the subject excreta, with strong probability.
We presented several models to uncover whether the nature of the common sharing that exists between injirleey, subject excreta is fulfilled by any of the given possibilities. In order to test the existing relationship between the samples, we tested the following scenarios:
s0 = common ancestor of two branches s1, s2, where s1 =/= s2
s0 -> s1 - s2
s0 = s1 = s2
s0 + s3 = s1 ; s0 -> s1
s0 = s3 = s2; s0 -> s1
With s0 = common node of branches s1, s2, and s3, with s1 =/= s2, s3, and s2 =/= s1, s3
Here's a few depicted relationships being presented below:
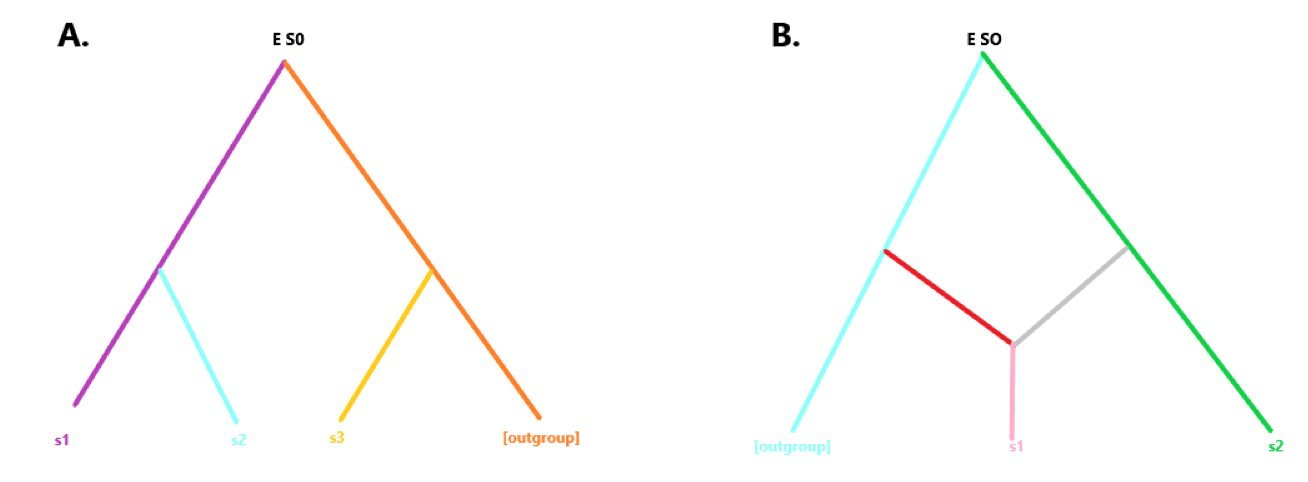
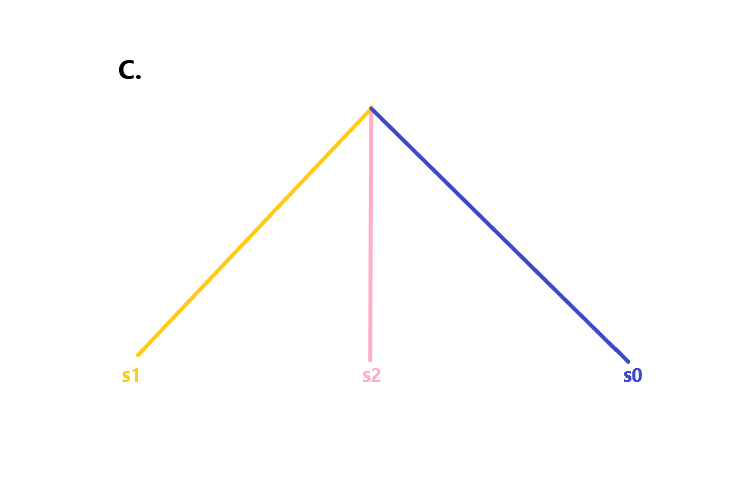
We found strong in support of unexpectedly high relation between the tested pair, but due to the very high similarity, were unable to favor one relationship over the other.
After this, we calculated F4 values with waaqo, subject excreta, injirleey, mbuti, in which we consistently saw significantly >0 values. supporting a vary high relatedness between injirleey and waaqo and or subject excreta.
In order to further investigate this, using HAPMIX, we establish that injirleey and waaqo are near identical at all tested loci, and form an exclusive sampled pair with few differentiating alleles, all of whom which are clustered, save a few, in stretches with known high mutational activity.
With this, we set out to measure whether the mutational history differentiating the near identical pair retroactively fits with the timeline of waaqo, the subject excreta age, and injirleey, by not only calculating the period elapsed between the samples, but by carbon14 dating the subject excreta.
Our C^14 dates for the subject excreta are 2 weeks +/- 1 week, well within the time interval in which waaqo has been banned and overlapping the date of the earliest reported sighting of similar excreta.
By calculating the mutation rate, with the formulae 0.5×10−9 basepair/yr, we arrive at roughly a 4 month time difference between the last known activity for waaqo and subject excreta samples, perfectly matching the C^14 dating of the subject excreta and the recorded date of sampling of the waaqo sample.
With this, we can state that the previously banned waaqo's dna was found to be present amongst the recently collected community samples, suggesting the banned member has indeed returned, but we have as yet have not established the true relation waaqo and injirleey may have with the subject excreta.
In order to gain a higher degree of confidence in discriminating power, we pooled together both waaqo and injirleey samples, and in turn ran them against the subject excreta with various outgroups to ascertian any special association between the two.
We then tested a series of private mutations found exclusively within the concerned samples, and by using the Hardy-Weinberg equation, tried to find the probability of identical frequencies, cross-referenced with data of probability of IBD of varying allele frequencies under various selection pressures.
a2 + 2ab + b2 = 1
a = 100, b = 0
.1002 + 2(.100 x 0) + 02
As we expected, the conditions of random mating are not satisfied and indicate either very high inbreeding, or that the same reoccuring sample is being tested here.
To test for further evidence of shared drift versus various outgroups, we used F3 and further F-stats to investigate the strength of any existing relationship between the various sets comprising injirleey, subject excreta, and waaqo.
We used F2 to calculate any exclusive drift between any two pair,
F2 (waaqo, injirleey), F2 (subject excreta, waaqo), F2 (subject excreta, waaqo)
Indeed we find not further evidence of near identicalness of drift between all sets, well beyond any expected IBD of two seperate persons. So much so in fact, that we can instead employ F2 as indicating allele differences in a point of time 1 and time 2 as expressed below
F2 = (pt1, pt2)
[CONTINUED BELOW]
Somalispot Department of Suugo Science
Ministry of Dila, Hala Dilo, Somaali Ma Aha
@Apollo Medical School, Institute for Cilmi Bariis
For weeks, the Somalispot community has been plagued with a trail of unfettered excreta of unidentified provenance coursing it's way through the sub-forum and forum at large. Several members of the community have taken at attempts in trying to source the origination of the perceivingly reproductive excreta. The characteristics of the subject excreta included, among several, a malodorous smell, elevated levels of hemoglobin blood iron, and plasma seemingly sourced from a variety of ruminants, and a tendency to proliferate due to unknown and yet uninvestigated means, and a symbiotic relationship with lice larvae and colonia.
The unbefore heard of set of traits that characterize the subject excreta have led to an abundance of theories to explain its source of origination. The following are a compilation of multiple theories put forth by several individuals and community members :
1) the Italian origination theory, which posits its substance on the rather unique Italian sourced name used by a user who is believed by some to be well timed and placed in association with this phenomena, going off the connection between Farabutto -> Farabuuto. This is despite it's usage amongst few Somalis.
2) the Eritrean origination theory, due to a battery if 15N and 13C- isotope analysis and dietary values of the subject excreta all aligning with the Tigray serie, consistent with its latent opposition to Amara and Galla sample sets, and the results of mass spectrometry having a positive association with previous collected Eritrean results
3) the Waaqo theory, due to the subjects excreta rather vague resemblant character traits
In addition to these three, there exists a forth theory, the hybrid theory, which characterized by the idea in which theories 2 and 3 having to not be mutually exclusive theories, but are in fact evidence of each other, based off a premise that waaqo was Eritrean, and not a non-Eritrean injirleey.
As of yet, the subject excreta has yet to have been identified with any association with Somalis, but by testing the probable occurrence of the latter theory, we found
O (x | y), with O (waaqo) = .30, and
O (waaqo | similarity of posts ) = .100
Which supports the Waaqo theory as having a higher probability of occurring with the known and considered probability factors.
In combination with our 13^C and 15^N isotope analysis, mass spectrometry, and dietary values all suggesting an Eritrean origin for the originating source of the subject excreta, independent of genetic evidence, we suggest with strong confidence in the hybrid theory, and for an Eritrean background for waaqoofpunt, and waaqoofpunt as the source of the offending subject excreta.
But due to the unsatisfactory and lack of robustness of the evidence thus given, the rather harsh argumentative and currently subjective nature of the discussion, and the uncooperative and fragile nature of the subject excreta, we found it necessary to perform a genetic analysis.
We genotyped samples which were collected with the extraction of a acidic core located on a remnant stain of the subject excreta which was received from @embarassing. We then prepared the sampled core, and successfully searched and extracted biopolymers, and using an extraction buffer consisting of proteinase k and EDTA and any needed decalsification, we estimated a yield of near 100% after centrifuge, and the ensuing yield was put in a polymerase chain reaction with several identical copies. We used capillary electrophoresis in the separation process, and identified all with a high power of discrimination all locus of interest in regards to the subject excreta. The MWF of our yield had a A260-80 ratio 1.9. We genotyped a total of 500k SNPs, and compared them to collated set of genotyped excreta remains and a collection of various African populations, with Han, Maya, Injirleey, san-Gaab, and Somali results, the latter and injirleey samples were collected being local community members. In order to test for the waaqo theory, we asked and received excrement collected and stored by the administration for purposes of identification of the two time banned member @waaqoofpunt. We similarly received further samples provided by community members of the same purported user found to be on twitter. We then pruned the collected set for any significant outliers that could hamper and mischarecterize our analysis, and had to remove the following samples for the following reasons:
@CaliTedesse (self identified Arabian, excess Somali ancestry)
@Samaalic Era (extremely high RoH)
@Medulla (unstable and erratic mutation rate).
@Basra (self identified as female, genotyped as XY)
We collated and integrated the Somali samples into a single high density sample in order to have a very high level of snp recall >99%.
By F-stats, we found a very high allele sharing between the subject excreta and the sole injirleey, and running F4 and F3 with Han, san-Gaab, and Chimp outgroup, see strong support for a common node including both injirleey and the subject excreta.
F4 was run as given: F4 (injirleey, subjectexcreta | san-Gaab, Han) = 0
thus supporting a common node and subset including injirleey, subjectexcreta.
We then ran F4 I with Somali, sanGaab to see a further common branching for this node
F4 (injirleey, subjectexcreta | Somali, sanGaab) was null, which establishes a very strong basis for a special relationship of injirleey and the subject excreta, with strong probability.
We presented several models to uncover whether the nature of the common sharing that exists between injirleey, subject excreta is fulfilled by any of the given possibilities. In order to test the existing relationship between the samples, we tested the following scenarios:
s0 = common ancestor of two branches s1, s2, where s1 =/= s2
s0 -> s1 - s2
s0 = s1 = s2
s0 + s3 = s1 ; s0 -> s1
s0 = s3 = s2; s0 -> s1
With s0 = common node of branches s1, s2, and s3, with s1 =/= s2, s3, and s2 =/= s1, s3
Here's a few depicted relationships being presented below:
We found strong in support of unexpectedly high relation between the tested pair, but due to the very high similarity, were unable to favor one relationship over the other.
After this, we calculated F4 values with waaqo, subject excreta, injirleey, mbuti, in which we consistently saw significantly >0 values. supporting a vary high relatedness between injirleey and waaqo and or subject excreta.
In order to further investigate this, using HAPMIX, we establish that injirleey and waaqo are near identical at all tested loci, and form an exclusive sampled pair with few differentiating alleles, all of whom which are clustered, save a few, in stretches with known high mutational activity.
With this, we set out to measure whether the mutational history differentiating the near identical pair retroactively fits with the timeline of waaqo, the subject excreta age, and injirleey, by not only calculating the period elapsed between the samples, but by carbon14 dating the subject excreta.
Our C^14 dates for the subject excreta are 2 weeks +/- 1 week, well within the time interval in which waaqo has been banned and overlapping the date of the earliest reported sighting of similar excreta.
By calculating the mutation rate, with the formulae 0.5×10−9 basepair/yr, we arrive at roughly a 4 month time difference between the last known activity for waaqo and subject excreta samples, perfectly matching the C^14 dating of the subject excreta and the recorded date of sampling of the waaqo sample.
With this, we can state that the previously banned waaqo's dna was found to be present amongst the recently collected community samples, suggesting the banned member has indeed returned, but we have as yet have not established the true relation waaqo and injirleey may have with the subject excreta.
In order to gain a higher degree of confidence in discriminating power, we pooled together both waaqo and injirleey samples, and in turn ran them against the subject excreta with various outgroups to ascertian any special association between the two.
We then tested a series of private mutations found exclusively within the concerned samples, and by using the Hardy-Weinberg equation, tried to find the probability of identical frequencies, cross-referenced with data of probability of IBD of varying allele frequencies under various selection pressures.
a2 + 2ab + b2 = 1
a = 100, b = 0
.1002 + 2(.100 x 0) + 02
As we expected, the conditions of random mating are not satisfied and indicate either very high inbreeding, or that the same reoccuring sample is being tested here.
To test for further evidence of shared drift versus various outgroups, we used F3 and further F-stats to investigate the strength of any existing relationship between the various sets comprising injirleey, subject excreta, and waaqo.
We used F2 to calculate any exclusive drift between any two pair,
F2 (waaqo, injirleey), F2 (subject excreta, waaqo), F2 (subject excreta, waaqo)
Indeed we find not further evidence of near identicalness of drift between all sets, well beyond any expected IBD of two seperate persons. So much so in fact, that we can instead employ F2 as indicating allele differences in a point of time 1 and time 2 as expressed below
F2 = (pt1, pt2)
[CONTINUED BELOW]










