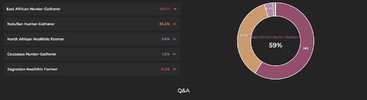
Basically this run you just did was a more a more refined version of the wheel chart they did for your stone age HG and farmer ancestry.okay
View attachment 273625
View attachment 273626
can you explain this and why are we using meheri yemeni and wtf is eth_mota
The ancient Egyptian you were getting was most likely just Arabian ancestry since they're both very similar.
Mota is the east african hunter-gatherer in your wheel chart, the thing is illustrative DNA only uses that as a proxy for your sub-Saharan ancestry but there's also a proto-Nilotic component. But they leave that out because there's no ancient sample to use. So we use Dinka as a close enough proxy to represent that.
For a better visualization of the time periods using a Cushitic reference do another RUN with this source
You don't have permission to view the spoiler content.
Log in or register now.