Apollo
VIP
@Grant
The Hadza and Sandawe are not Khoisan. They are proto-Omotic. There are no Khoisan in East Africa. See this study:
https://academic.oup.com/gbe/article/10/3/875/4935243
We have performed genetic analyses to better understand the history of the Hadza and Sandawe populations in Tanzania. Using a combination of semi-supervised and unsupervised clustering analysis with a large global reference panel, we better defined ancestral composition. In the context of the 19 ancestries we previously detected (Shriner et al. 2014), we found that the Hadza and Sandawe populations shared a distinct ancestry that we eponymously named Hadza ancestry (because six Hadza individuals were homogeneous for this ancestry). We also found that genotype and sequence data support an early divergence model for Hadza ancestry.
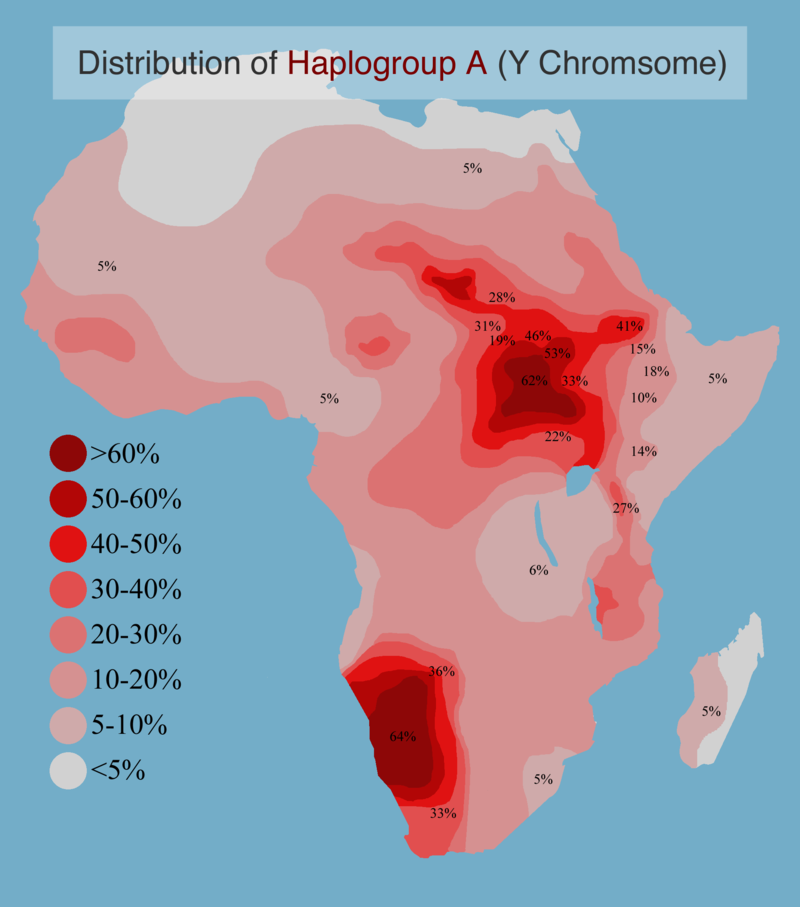
We detected low levels of mixed ancestry among a subset of Hadza individuals. Specifically, we detected Niger-Congo ancestry in 6 of 13 Hadza individuals. We also detected Cushitic ancestry in 3 of 13 Hadza individuals and at lower levels than Niger-Congo ancestry. Additionally, we detected Nilo-Saharan ancestry in one Hadza individual. These results are consistent with the presence of both E1b1a Y chromosome haplogroups, common in populations with Niger-Congo ancestry, and E1b1b Y chromosome haplogroups, common in populations with Nilo-Saharan and Cushitic ancestries, in the Hadza population (Tishkoff et al. 2007). Collectively, the autosomal data and Y chromosome data provide evidence for the presence of Hadza, Niger-Congo, Cushitic, and Nilo-Saharan ancestry in the Hadza population. Within the Hadza sample, the simultaneous presence of ancestrally heterogeneous individuals with large amounts of Niger-Congo ancestry and individuals homogeneous for Hadza ancestry is consistent with very recent admixture.
We detected a more complex mixture of ancestries in the Sandawe individuals than in the Hadza individuals. Whether this finding reflects more inter-mating in the Sandawe population or more loss of lineages in the Hadza population is unknown. We identified Niger-Congo (more specifically, eastern and southern Bantu-speaking) ancestry, Cushitic ancestry, Omotic ancestry, Hadza ancestry, and Khoisan ancestry in all the Sandawe individuals. Additionally, we identified Arabian ancestry and eastern Pygmy ancestry in a minority of Sandawe individuals. Compared with the Y chromosomal haplogroup frequencies in the Hadza population, the Sandawe population has more E1b1a and E1b1b and less B2 (Tishkoff et al. 2007), consistent with our autosomal findings of higher amounts of Niger-Congo and Cushitic ancestry in the Sandawe individuals.
Tishkoff et al. (2009) reported Hadza and Sandawe ancestries but did not include samples of speakers of Omotic languages. Our results are consistent with Hadza ancestry having formed by a splitting process. In contrast, our results indicate that Sandawe ancestry reflects a mixture of eastern and southern African Bantu-speaking, Cushitic, Omotic, Hadza, and Khoisan ancestries.
Pickrell et al. (2012) inferred the presence in both the Hadza and Sandawe populations of ancestry shared with Khoisan-speaking peoples. Based on the clustering analyses, we found no Khoisan ancestry in the Hadza individuals and low levels of Hadza, Khoisan, and Omotic ancestries in the Sandawe individuals. However, analysis of ancestral allele frequencies revealed a migration event between Khoisan and Hadza ancestries and a migration event between Khoisan and Omotic ancestries. Thus, ancestry in the Hadza and Sandawe populations shared with Khoisan-speaking populations could reflect the distant common ancestor of Hadza, Khoisan, and Omotic ancestries or these more recent migration events.
Mitochondrial DNA provides uniparental information about maternal lineages. The divergence of L0d and L0a’b’f’k haplogroups occurred ∼119,000 [100,100–138,200] years ago, the divergence of L0k and L0a’b’f haplogroups occurred ∼98,700 years [82,300 to 115,400] ago, and the divergence of L0a occurred ∼42,400 [33,000–52,000] years ago (Rito et al. 2013). The! Xun have >50% L0d and ∼25% L0k, whereas the Hadza population has 5% L0a and the Sandawe population has 26% L0a, L0d, and L0f (Tishkoff et al. 2007). The L0k haplogroup was not observed in either the Hadza or Sandawe populations (Tishkoff et al. 2007). These data are consistent with the early divergence of Hadza ancestry, that is, before the divergence of L0k, and a more recent acquisition of L0a. However, the absence of L0k could have resulted from loss in the Hadza and Sandawe populations due to random genetic drift. Y chromosome DNA provides uniparental information about paternal lineages. The emergence of B-M181 105,800 years ago and B2-M182 100,600 years ago (Poznik et al. 2016) are consistent with an early divergence of Hadza ancestry.
Collectively, autosomal, Y, and mitochondrial DNA support early divergence of Hadza ancestry (Knight et al. 2003; Tishkoff et al. 2007). Lachance et al.’s (2012) conclusion of late divergence was based on a neighbor-joining tree; the assumption of treeness or bifurcation is violated by admixture and gene flow, thus invalidating their conclusion. An early divergence of Hadza and Khoe-San peoples is consistent with the grouping of Hadza and Khoisan languages (Knight et al. 2003). On the other hand, evidence for gene flow between 7.5 and 20 thousand years ago is consistent with the hypothesis that click sounds are a recent addition to Hadza and Sandawe languages (Rito et al. 2013). Our results suggest a third possibility. The semi-supervised analysis revealed that Hadza ancestry is closer to Omotic ancestry than to Khoisan ancestry. Also, Omotic ancestry does not cluster with Arabian, Berber, or Cushitic ancestries, consistent with the hypothesis that Omotic languages are not part of the Afroasiatic language family (Theil 2006). Taken together, our genetic findings support a phylolinguistic hypothesis that Omotic and Hadza languages form a language family (Elderkin 1982). Furthermore, if both Cushitic and Niger-Congo ancestries in the Sandawe sample are comparatively recently acquired, then the core of the Sandawe sample is predominantly Omotic, supporting a phylolinguistic hypothesis that the Sandawe language also belongs with Omotic and Hadza languages. The hypothesis that Hadza and Sandawe peoples are not Khoe-San peoples is supported by previous osteological and serogenetic studies (Morris 2002).
[....]
Finally, our ancestry analyses support the hypothesis that Omotic, Hadza, and Sandawe languages group together, rather than Omotic languages belonging to the Afroasiatic family and Hadza and Sandawe languages belonging to the Khoisan family.
https://academic.oup.com/gbe/article/10/3/875/4935243
Keep in mind East Somalis carry 0% ancestry from these groups.
The Hadza and Sandawe are not Khoisan. They are proto-Omotic. There are no Khoisan in East Africa. See this study:
https://academic.oup.com/gbe/article/10/3/875/4935243
We have performed genetic analyses to better understand the history of the Hadza and Sandawe populations in Tanzania. Using a combination of semi-supervised and unsupervised clustering analysis with a large global reference panel, we better defined ancestral composition. In the context of the 19 ancestries we previously detected (Shriner et al. 2014), we found that the Hadza and Sandawe populations shared a distinct ancestry that we eponymously named Hadza ancestry (because six Hadza individuals were homogeneous for this ancestry). We also found that genotype and sequence data support an early divergence model for Hadza ancestry.
We detected low levels of mixed ancestry among a subset of Hadza individuals. Specifically, we detected Niger-Congo ancestry in 6 of 13 Hadza individuals. We also detected Cushitic ancestry in 3 of 13 Hadza individuals and at lower levels than Niger-Congo ancestry. Additionally, we detected Nilo-Saharan ancestry in one Hadza individual. These results are consistent with the presence of both E1b1a Y chromosome haplogroups, common in populations with Niger-Congo ancestry, and E1b1b Y chromosome haplogroups, common in populations with Nilo-Saharan and Cushitic ancestries, in the Hadza population (Tishkoff et al. 2007). Collectively, the autosomal data and Y chromosome data provide evidence for the presence of Hadza, Niger-Congo, Cushitic, and Nilo-Saharan ancestry in the Hadza population. Within the Hadza sample, the simultaneous presence of ancestrally heterogeneous individuals with large amounts of Niger-Congo ancestry and individuals homogeneous for Hadza ancestry is consistent with very recent admixture.
We detected a more complex mixture of ancestries in the Sandawe individuals than in the Hadza individuals. Whether this finding reflects more inter-mating in the Sandawe population or more loss of lineages in the Hadza population is unknown. We identified Niger-Congo (more specifically, eastern and southern Bantu-speaking) ancestry, Cushitic ancestry, Omotic ancestry, Hadza ancestry, and Khoisan ancestry in all the Sandawe individuals. Additionally, we identified Arabian ancestry and eastern Pygmy ancestry in a minority of Sandawe individuals. Compared with the Y chromosomal haplogroup frequencies in the Hadza population, the Sandawe population has more E1b1a and E1b1b and less B2 (Tishkoff et al. 2007), consistent with our autosomal findings of higher amounts of Niger-Congo and Cushitic ancestry in the Sandawe individuals.
Tishkoff et al. (2009) reported Hadza and Sandawe ancestries but did not include samples of speakers of Omotic languages. Our results are consistent with Hadza ancestry having formed by a splitting process. In contrast, our results indicate that Sandawe ancestry reflects a mixture of eastern and southern African Bantu-speaking, Cushitic, Omotic, Hadza, and Khoisan ancestries.
Pickrell et al. (2012) inferred the presence in both the Hadza and Sandawe populations of ancestry shared with Khoisan-speaking peoples. Based on the clustering analyses, we found no Khoisan ancestry in the Hadza individuals and low levels of Hadza, Khoisan, and Omotic ancestries in the Sandawe individuals. However, analysis of ancestral allele frequencies revealed a migration event between Khoisan and Hadza ancestries and a migration event between Khoisan and Omotic ancestries. Thus, ancestry in the Hadza and Sandawe populations shared with Khoisan-speaking populations could reflect the distant common ancestor of Hadza, Khoisan, and Omotic ancestries or these more recent migration events.
Mitochondrial DNA provides uniparental information about maternal lineages. The divergence of L0d and L0a’b’f’k haplogroups occurred ∼119,000 [100,100–138,200] years ago, the divergence of L0k and L0a’b’f haplogroups occurred ∼98,700 years [82,300 to 115,400] ago, and the divergence of L0a occurred ∼42,400 [33,000–52,000] years ago (Rito et al. 2013). The! Xun have >50% L0d and ∼25% L0k, whereas the Hadza population has 5% L0a and the Sandawe population has 26% L0a, L0d, and L0f (Tishkoff et al. 2007). The L0k haplogroup was not observed in either the Hadza or Sandawe populations (Tishkoff et al. 2007). These data are consistent with the early divergence of Hadza ancestry, that is, before the divergence of L0k, and a more recent acquisition of L0a. However, the absence of L0k could have resulted from loss in the Hadza and Sandawe populations due to random genetic drift. Y chromosome DNA provides uniparental information about paternal lineages. The emergence of B-M181 105,800 years ago and B2-M182 100,600 years ago (Poznik et al. 2016) are consistent with an early divergence of Hadza ancestry.
Collectively, autosomal, Y, and mitochondrial DNA support early divergence of Hadza ancestry (Knight et al. 2003; Tishkoff et al. 2007). Lachance et al.’s (2012) conclusion of late divergence was based on a neighbor-joining tree; the assumption of treeness or bifurcation is violated by admixture and gene flow, thus invalidating their conclusion. An early divergence of Hadza and Khoe-San peoples is consistent with the grouping of Hadza and Khoisan languages (Knight et al. 2003). On the other hand, evidence for gene flow between 7.5 and 20 thousand years ago is consistent with the hypothesis that click sounds are a recent addition to Hadza and Sandawe languages (Rito et al. 2013). Our results suggest a third possibility. The semi-supervised analysis revealed that Hadza ancestry is closer to Omotic ancestry than to Khoisan ancestry. Also, Omotic ancestry does not cluster with Arabian, Berber, or Cushitic ancestries, consistent with the hypothesis that Omotic languages are not part of the Afroasiatic language family (Theil 2006). Taken together, our genetic findings support a phylolinguistic hypothesis that Omotic and Hadza languages form a language family (Elderkin 1982). Furthermore, if both Cushitic and Niger-Congo ancestries in the Sandawe sample are comparatively recently acquired, then the core of the Sandawe sample is predominantly Omotic, supporting a phylolinguistic hypothesis that the Sandawe language also belongs with Omotic and Hadza languages. The hypothesis that Hadza and Sandawe peoples are not Khoe-San peoples is supported by previous osteological and serogenetic studies (Morris 2002).
[....]
Finally, our ancestry analyses support the hypothesis that Omotic, Hadza, and Sandawe languages group together, rather than Omotic languages belonging to the Afroasiatic family and Hadza and Sandawe languages belonging to the Khoisan family.
https://academic.oup.com/gbe/article/10/3/875/4935243
Keep in mind East Somalis carry 0% ancestry from these groups.